
Patrick Hsu: A New DNA Editing Technology Reported In Science Magazine
Patrick Hsu, Co-Founder of Arc Institute, posted on LinkedIn:
”Today in Science Magazine, we report a new DNA editing technology to seamlessly write massive changes into the right place in the human genome.
The reason gene editing hasn’t transformed human health is that current gene editing technologies like CRISPR are very limited.
The problem with CRISPR is that it cuts up your DNA, and then hopes that unreliable cellular DNA repair will make the wanted edit.
famously called it genome vandalism. More precise versions of CRISPR only edit less than 100 bases – often only a single base. Therefore, it’s not suited to make large changes safely.
However, most diseases are not the result of mutations in one location. Instead, their causes are spread all across the 3 billion base pairs in the genome.
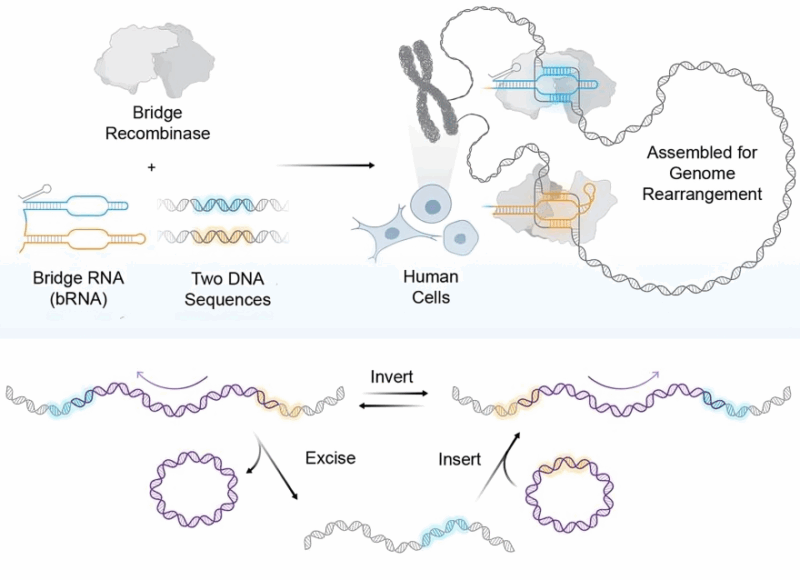
We found bridge RNAs in bacterial “jumping genes” that allow us to make safe and arbitrary changes (insert, cut out, or flip) to every nucleotide within (up to) a 1 million bp sequence in your DNA.
In the paper, we show that we can correct the disease-causing DNA repeats that cause Friedreich’s ataxia (which is a rare neurological disease). The same approach could be applied to Huntington’s and other repeat expansion disorders.
This work was a wonderful collaboration with my Arc Institute cofounder Silvana Konermann and led by the indefatigable Nicholas Perry, alongside our amazing bridge editing team: Liam Bartie, Dhruva Katrekar, Gabriel Gonzalez, Matthew Durrant, James Pai, Alison Fanton, Juliana Martins, Masahiro Hiraizumi, Chiara Ricci-Tam, and Hiroshi Nishimasu”
Read the full article here.
Title: Megabase-scale human genome rearrangement with programmable bridge recombinases
Authors: Nicholas T. Perry, Liam J. Bartie, Dhruva Katrekar, Gabriel A. Gonzalez, Matthew G. Durrant, James J. Pai, Alison Fanton, Juliana Q. Martins, Masahiro Hiraizumi, Chiara Ricci-Tam, Hiroshi Nishimasu, Silvana Konermann, Patrick D. Hsu
Stay updated with Hemostasis Today.
-
Feb 26, 2026, 15:58Daniel Victor Ortigoza։ Lipoprotein(a) Levels Predict Long-Term Cardiovascular Risk in Women
-
Feb 26, 2026, 15:51Augustina Isioma Ikusemoro: The Real Miracle in Trauma Care Happens Behind the Scenes
-
Feb 26, 2026, 15:42Jim Hoffman։ Targeting NETosis to Improve Perfusion and Reduce Thrombosis in AMI
-
Feb 26, 2026, 15:40Elvira Grandone: ISTH Launches An International Survey to Capture Real-World Practice of Pregnancy Anemia
-
Feb 26, 2026, 15:39Ashok Yadav: Placental Glycogen as a Vital Energy Reserve for Fetal and Placental Health
-
Feb 26, 2026, 15:37Nayab Ahmed: Main Reasons for Washing Platelet-Rich Plasma
-
Feb 26, 2026, 15:36Robert Negrin Shares His Outlook for the Year Ahead as ASH President
-
Feb 26, 2026, 15:24Nancy Shapiro: Excited to See This Publication on Current Landscape of Anticoagulation Stewardship Released
-
Feb 26, 2026, 15:15Ahmed Nasreldein: Stay Current on Key Updates in Ischemic and Hemorrhagic Stroke

