
New Research on CRISPR’s Hidden Risks: Reviewed by Wolfgang Miesbach
Wolfgang Miesbach, Professor of Medicine at Frankfurt University Hospital, shared a proud post on Linkedin:
“CRISPR’s hidden danger: New Nature Communication from genome editing safety experts Toni Cathomen, Carla Fuster García and Clotilde Aussel reveals concerns about ‘precision’ enhancers!
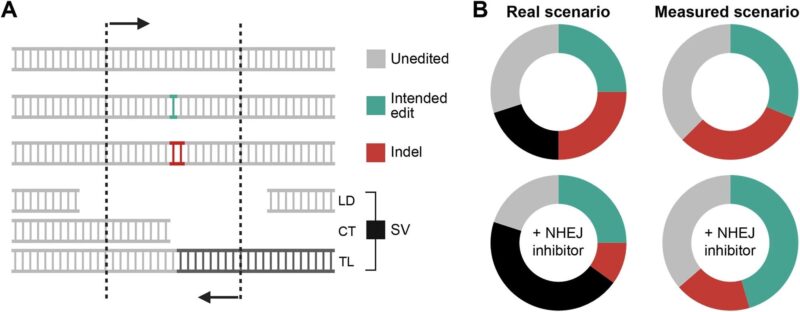
Beyond Off-Target Effects: A Worse Problem: The field has been focused on off-target effects (off-target sites prediction, and off-target activity measurement)- where CRISPR cuts at unintended sites with similar sequences to the target.
But this study reveals something potentially worse: structural variations (SVs) including massive chromosomal chaos that happens even at the intended target site.
DNA-PKcs inhibitors like AZD7648 – compounds used to make CRISPR more precise by promoting homology-directed repair (HDR) over error-prone repair – cause a 1000-fold increase in chromosomal translocations.
The Catastrophic Outcomes:
- Megabase-scale deletions spanning millions of DNA bases
- 47.8% of cells losing entire chromosome arms
- Kilobase-scale deletions increased up to 35-fold
- Chromosomal translocations between different chromosomes
Chromothripsis – catastrophic chromosome shattering
Clinical Reality Check:
Casgevy – the first approved CRISPR therapy – targets BCL11A for sickle cell disease.
BCL11A editing frequently causes large kilobase-scale deletions in stem cells.
8% of African ancestry patients carry variants creating additional off-target sites.
Repair Pathway Problem:
In human cells, there are two main DNA repair pathways after CRISPR cuts:
- NHEJ (Non-Homologous End Joining) fast but error-prone
- HDR (Homology-Directed Repair) precise but inefficient
By inhibiting DNA-PKcs to boost HDR, researchers accidentally created genomic chaos.
Detection Crisis:
Standard sequencing completely misses these changes because large deletions remove PCR primer sites, making them “invisible” and leading to massive overestimation of success rates.
Սolutions From the Authors:
The authors discuss better detection methods (CAST-Seq, long-read sequencing), safer enhancement approaches(53BP1 inhibition), alternative strategies (base editors), and rethinking necessity of ultra-high efficiency.
Their conclusion: We need ՛holistic, treatment-centered evaluation of both off-target and aberrant on-target effects to ensure therapeutic efficacy is not achieved at the expense of unintended consequences՛”
Read the full article here.
Title: The hidden risks of CRISPR/Cas: structural variations and genome integrity
Authors: Clotilde Aussel, Toni Cathomen, Carla Fuster-García
Stay updated on the latest scientific advances with Hemostasis Today.
-
Feb 25, 2026, 13:36Zoltan Nagy: Antibody Response Can Shift from Recognizing the Adenoviral Protein to Targeting PF4 in Rare VITT Cases
-
Feb 25, 2026, 13:08Sam K. Saha: Same Clot Against Fast and Slow Systems
-
Feb 25, 2026, 13:04Haykaz Muradyan: The Use of REBOA in Obstetric Haemorrhage
-
Feb 25, 2026, 12:53Jan Hartmann: NexSys Plasma Collection System Receives FDA Clearance
-
Feb 25, 2026, 12:45Zain Khalpey: Secondary Stroke Prevention as The Most Important Opportunity to Reduce Disability
-
Feb 25, 2026, 12:40Jeff June: Stroke Care Is Clinically Mature, Stroke Biology Is Still Evolving
-
Feb 25, 2026, 12:36Wolfgang Miesbach: Repurposing Metformin – From Sugar Control to Thrombus Control
-
Feb 25, 2026, 12:30Bethany Brown: There Is A Need for A Validated Replacement for The Discontinued 2,3-DPG Assay
-
Feb 25, 2026, 12:20Register for A Webinar on von Willebrand Disease in Ageing – EHC

